Precision Oncology through Interpretable Machine Learning
Tumor vs Normal Cell Signatures from Single-Cell Data
🎯 Abstract & Objectives
This project analyzes single-cell transcriptomics data from human breast cancers to distinguish tumor and normal cell populations through comprehensive computational methods. Our workflow integrates data acquisition, feature engineering,oxphos, protooncogene categorisation, copy number variation analysis, machine learning classification, and therapeutic target identification using DGIdb, Open Targets databases and Clinical trial datasets.
Key Goals
- Feature Engineering: Extract biologically meaningful signatures from scRNA-seq data
- Machine Learning Model: Develop interpretable ML models to distinguish tumor vs normal cells
- Cross-Dataset Validation: Ensure model generalizability across multiple breast cancer datasets
- Therapeutic Discovery: Identify clinically relevant gene targets for precision oncology
Analysis Workflow Overview
📊 Datasets & Methodology
Single-Cell RNA-seq Datasets
- GSE176078: Primary training dataset - Breast cancer scRNA-seq
- GSE161529: Cross-Validation dataset - Additional breast cancer samples
- GSE180286: Cross-Validation dataset - Additional breast cancer samples
🧬 Feature Engineering
- Cell cycle scoring
- Apoptosis signatures
- Ribosomal content analysis
- OXPHOS pathway activity
- CNV burden quantification
🤖 Machine Learning
- XGBoost classification
- Threshold optimization
- Cross-validation
- SMOTE for class balancing
- SHAP for interpretability
🔬 CNV Analysis
- Chromosomal instability
- inferCNV pipeline
- Gencode v44 annotations
- Copy number scoring
💊 Therapeutic Mapping
- DGIdb druggability data
- Open Targets associations
- Pathway enrichment
- Clinical trial integration
Analysis Workflow
Data Loading
Feature Engineering
CNV Analysis
ML Classification
SHAP Analysis
Therapeutic Mapping
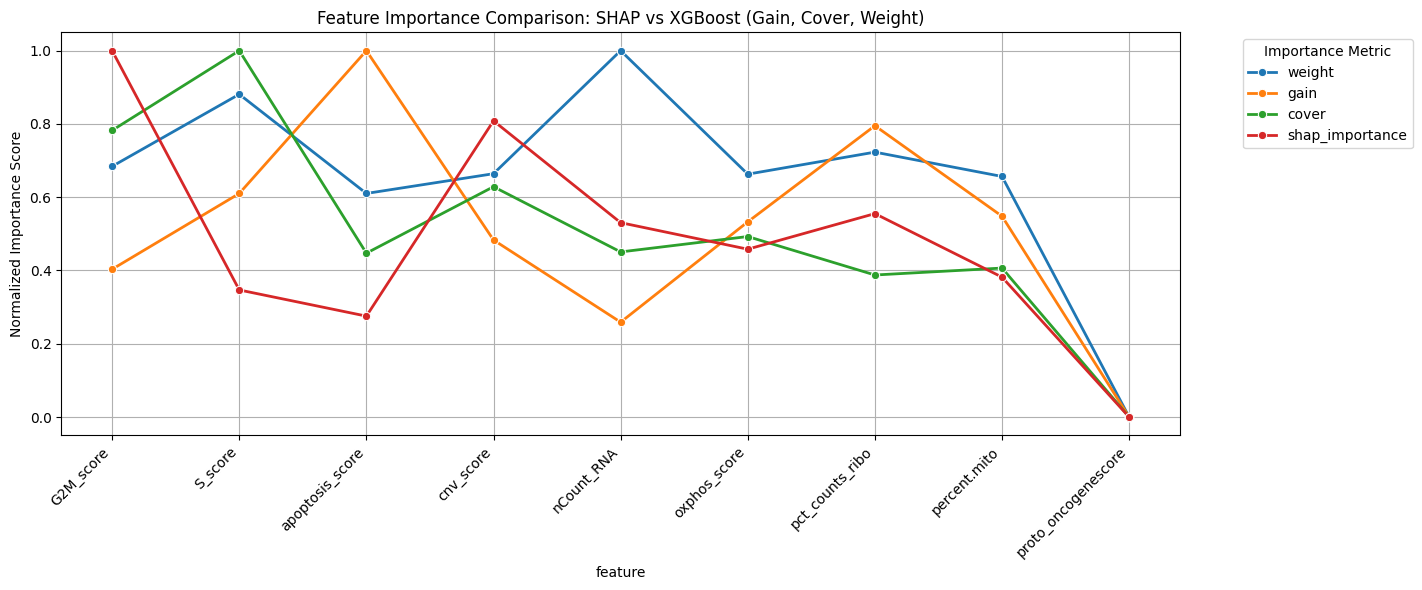
Feature Importance Analysis
📈 Key Results & Performance
🎯 Classification Performance
- AUC:89+ Excellent discrimination
- Precision: 0.95+ High accuracy
- Recall: 0.89+ Comprehensive detection
- Robust cross-dataset validation
🧬 Biological Insights
- CNV burden as key discriminator
- Cell cycle dysregulation in tumors
- Metabolic pathway differences
- Oncogene expression patterns
🔍 Feature Importance
- CNV Score: Top discriminative feature
- Cell Cycle: S/G2M phase enrichment
- OXPHOS: Metabolic reprogramming
- Ribosomal: Translation activity
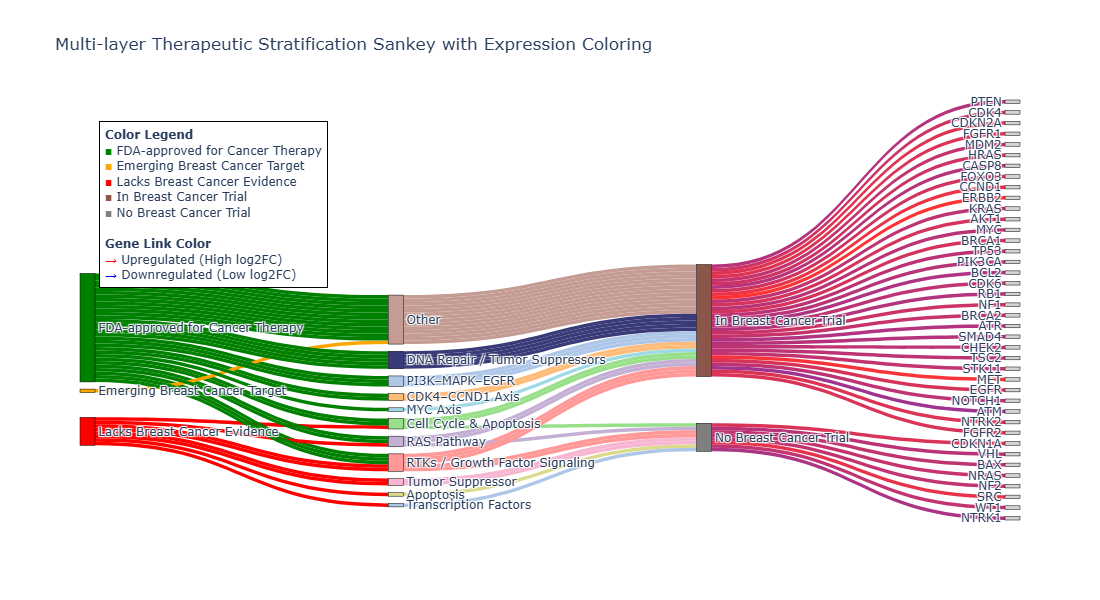
💊 Therapeutic Targets
- PI3K/MAPK/EGFR pathway genes
- CDK4–CDK6–CCND1 pathway genes
- Emerging and Underexplored targets
- DGIdb status, Open Target Score, Breast cancer support categorisation and Clinical trial mapping
🌟 Major Discoveries
- Universal Tumor Signatures: Identified robust features that discriminate tumor cells across multiple datasets
- CNV as Biomarker: Copy number variation burden emerged as the strongest predictor of malignancy
- Pathway Networks: Mapped therapeutic targets within key oncogenic pathways (PI3K, MAPK, EGFR)
- Clinical Relevance: Integrated druggability and clinical trial data for precision medicine applications
Model Performance Visualization
🎛️ Technical Implementation
🐍 Core Technologies
- Scanpy: Single-cell analysis framework
- InferCNVpy: Copy number variation detection
- XGBoost: Gradient boosting classification
- SHAP: Model interpretability and feature importance
- Pandas/NumPy: Data manipulation and analysis
- Matplotlib/Seaborn: Visualization and plotting
📊 Data Processing Pipeline
- Quality Control: Cell and gene filtering
- Normalization: Library size and log transformation
- Feature Selection: Highly variable genes identification
- Dimensionality Reduction: PCA and UMAP embedding
🔬 Validation Strategy
- Cross-Dataset Validation: Model trained on GSE176078, validated on GSE161529 & GSE180286
- Threshold Optimization: Fine-tuning model parameters for improved performance
- SHAP Analysis: Feature importance and model interpretability
- Biological Concordance: Pathway identification and literature concordance
Category Sankey Plot Analysis
🔮 Impact & Future Directions
🏥 Clinical Applications
- Therapeutic Stratification: Patient-specific treatment recommendations
- Drug Repositioning: Novel therapeutic applications for existing drugs
- Biomarker Discovery: CNV burden as prognostic indicator
🚀 Future Enhancements
- Multi-Modal Integration: Incorporate spatial, genomic, and proteomic data
- Real-Time Analysis: Deploy models for clinical decision support
- Pan-Cancer Extension: Generalize approach to other cancer types
‼️ Potential Challenges:
- Data Privacy: Ensuring patient confidentiality and data security
- Model Generalizability: Addressing biases in training data
- Clinical Validation: Rigorous testing in real-world settings